Nagahama
Contents
BioBrick Parts to achieve each medal requirement
We created a new BioBrick[1] for bronze and silver medal criterion. We have documented and submitted them to igem Registry. We introduce sod2 [http://parts.igem.org/Part:BBa_K2256000 (BBa_K2256000)]. This biobrick is Key BioBrics in our project. sod2 from Schizosaccharomyces pombe encodes a Na+/H+ antiporter enzyme, giving tolerance to high sodium and lithium concentrations. We used this part to give Saccharomyces cerevisiae salt tolerance. Insertion of sod2 gene (Na + / H + antiporter) into the plasmid and it's introduction enable to grow at high salt concentration.
Bronze
Confirmation whether sod2 were inserted into pSB1C3 and pYES2 plasmid.
The front and the back part of the intron of sod2 were separately amplified by PCR. The forward primer of sod2 located back the intron contained the sequence of reverse Primer in the front of intron as an adapter.
Using these two PCR products, fusion PCR was performed.
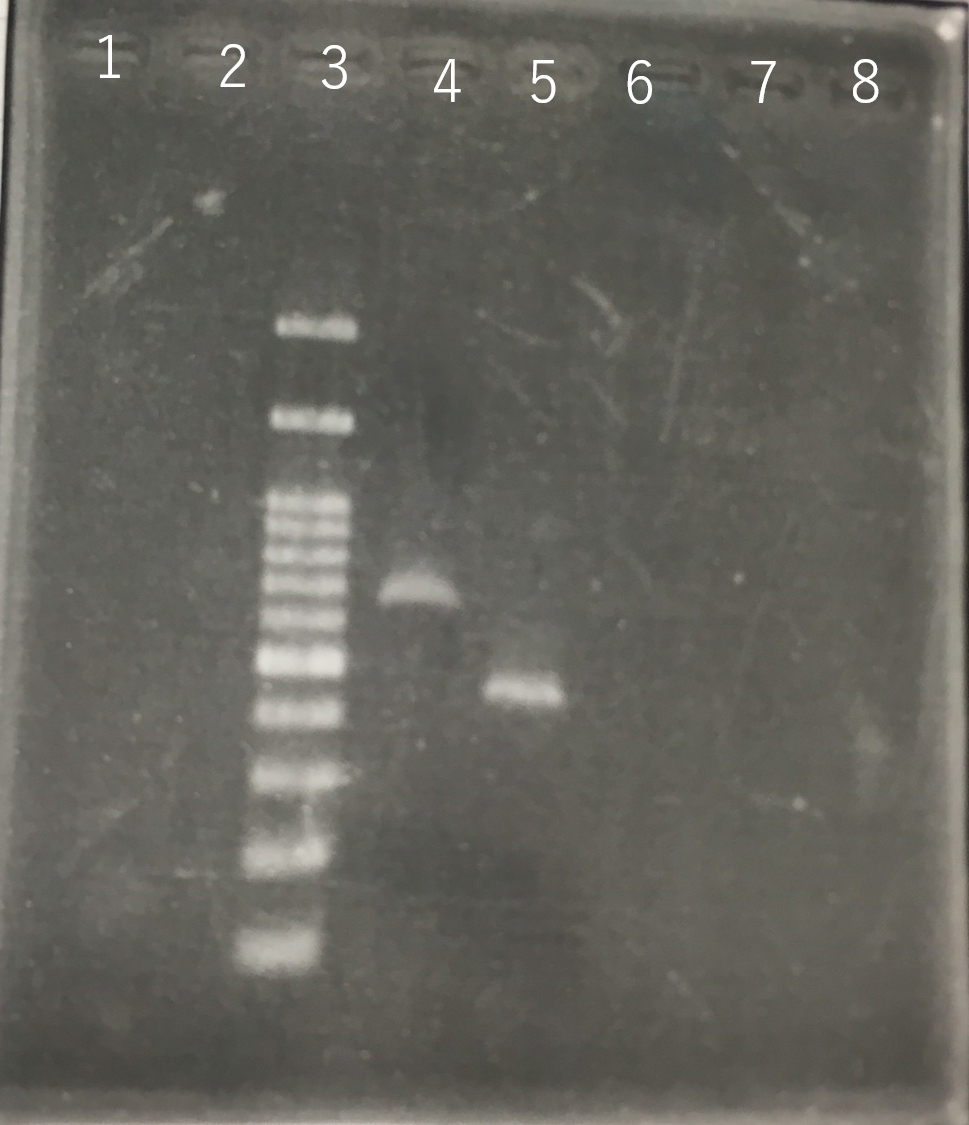
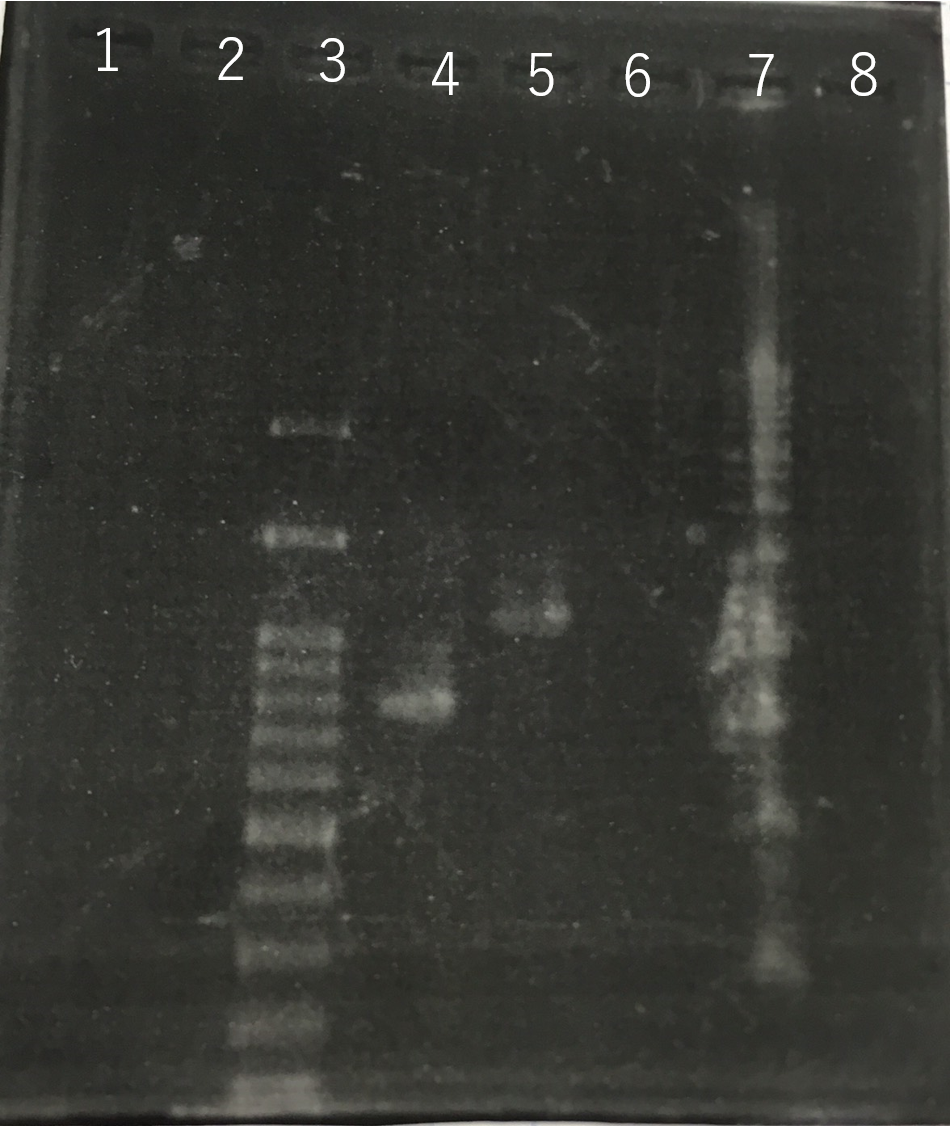
The completed PCR product of the coding region of sod2 was integrated into pSB1C3 and pYES2 plasmid and transfered the resultant plasmid into E.coli. Plasmid extraction was performed from the transformed E. coli . Using the restriction enzyme used to ligate the PCR product of sod 2 with each Plasmid, the plasmid extracted from E. coli was cut and the length was confirmed by electrophoresis.The length of pYES2 plasmid is 5856bp.The length of pSB1C3 is 2070bp.
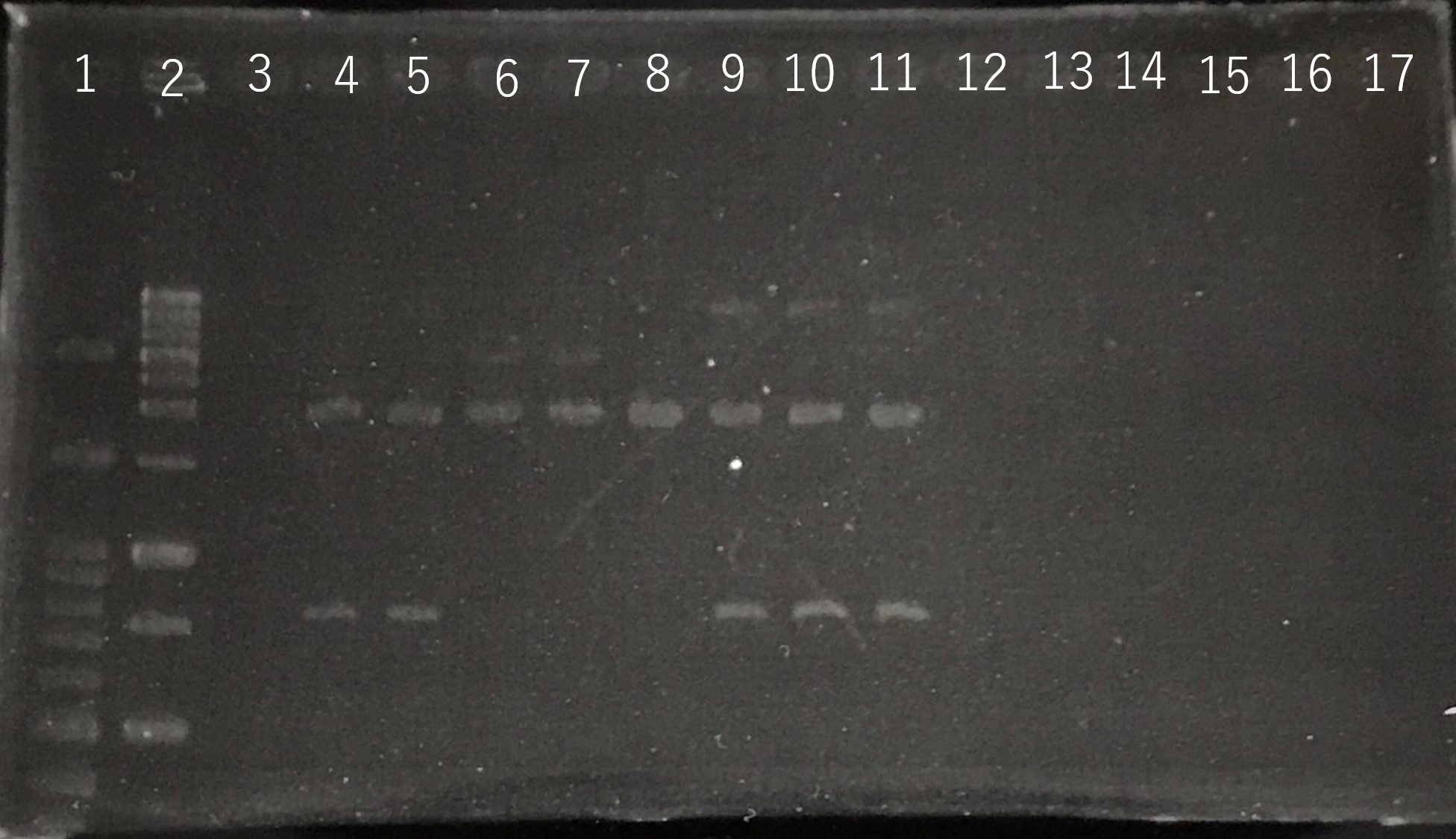
Lane1:100bp DNA ladder
Lane2:1kb DNA ladder
Lane3:Nothing
Lane4:Plasmid extraction from colony1
Lane5:Plasmid extraction from colony2
Lane6:Plasmid extraction from colony3
Lane7:Plasmid extraction from colony4
Lane8:Plasmid extraction from colony5
Lane9:Plasmid extraction from colony6
Lane10:Plasmid extraction from colony7
Lane11:Plasmid extraction from colony8
These result (fig.1,2,3,4) showed that sod2 was introduced into pSB1C3 extracted colony 4 and 5.
We have documented and submitted this plasmid as BioBrick Part [http://parts.igem.org/Part:BBa_K2256000 (BBa_K2256000)]. to iGEM Registry.
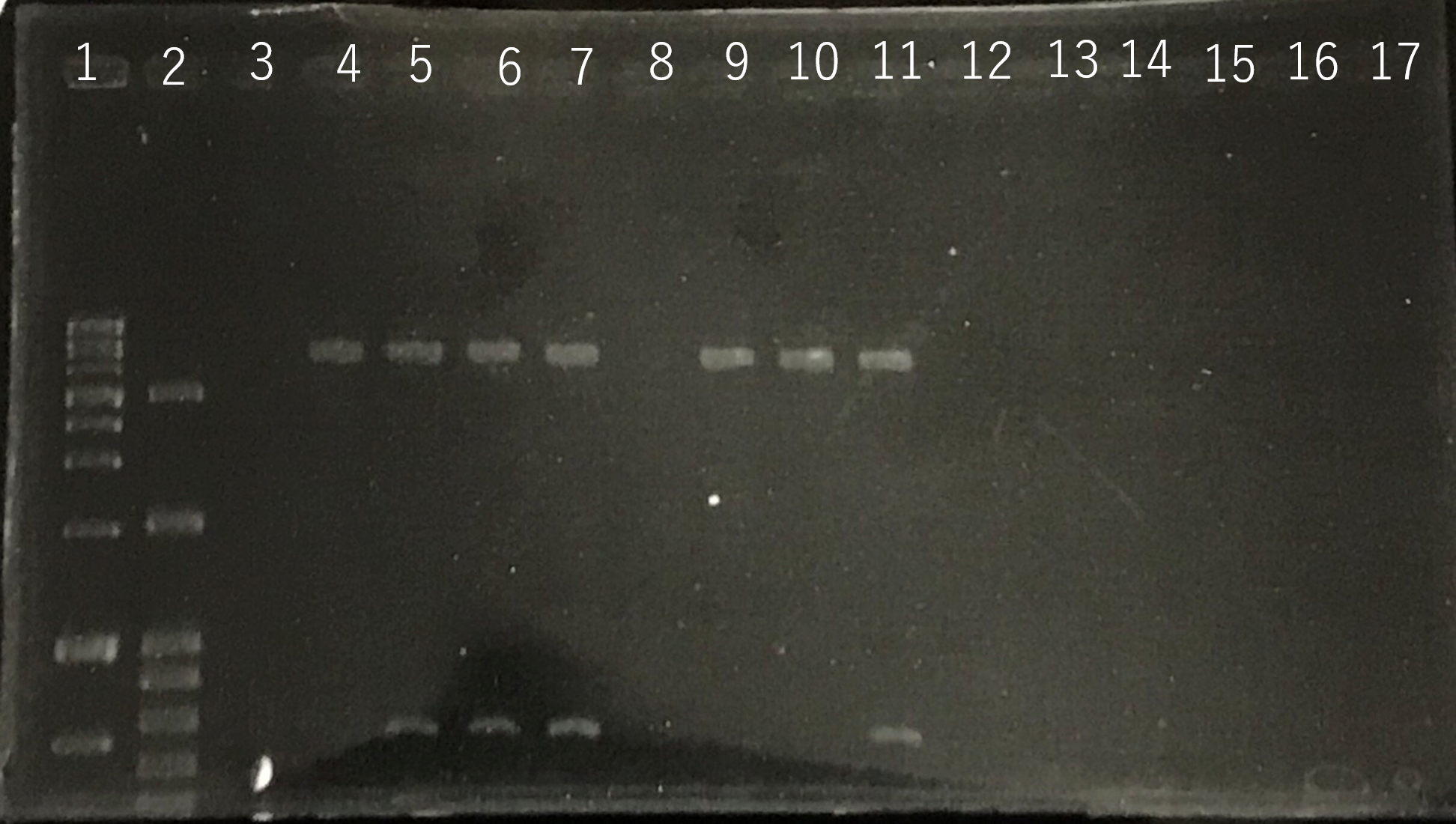
Lane1:1kb DNA ladder
Lane2:100bp DNA ladder
Lane3:Nothing
Lane4:Plasmid extraction from colony1
Lane5:Plasmid extraction from colony2
Lane6:Plasmid extraction from colony3
f Lane7:Plasmid extraction from colony4
Lane8:Plasmid extraction from colony5
Lane9:Plasmid extraction from colony6
Lane10:Plasmid extraction from colony7
Lane11:Plasmid extraction from colony8
These result (fig.1,2,3,5) showed that sod2 was introduced into pYES2 extracted colony 2, 3, 4 and 8. S. cerevisiae BY4742 was transformed with the plasmid.
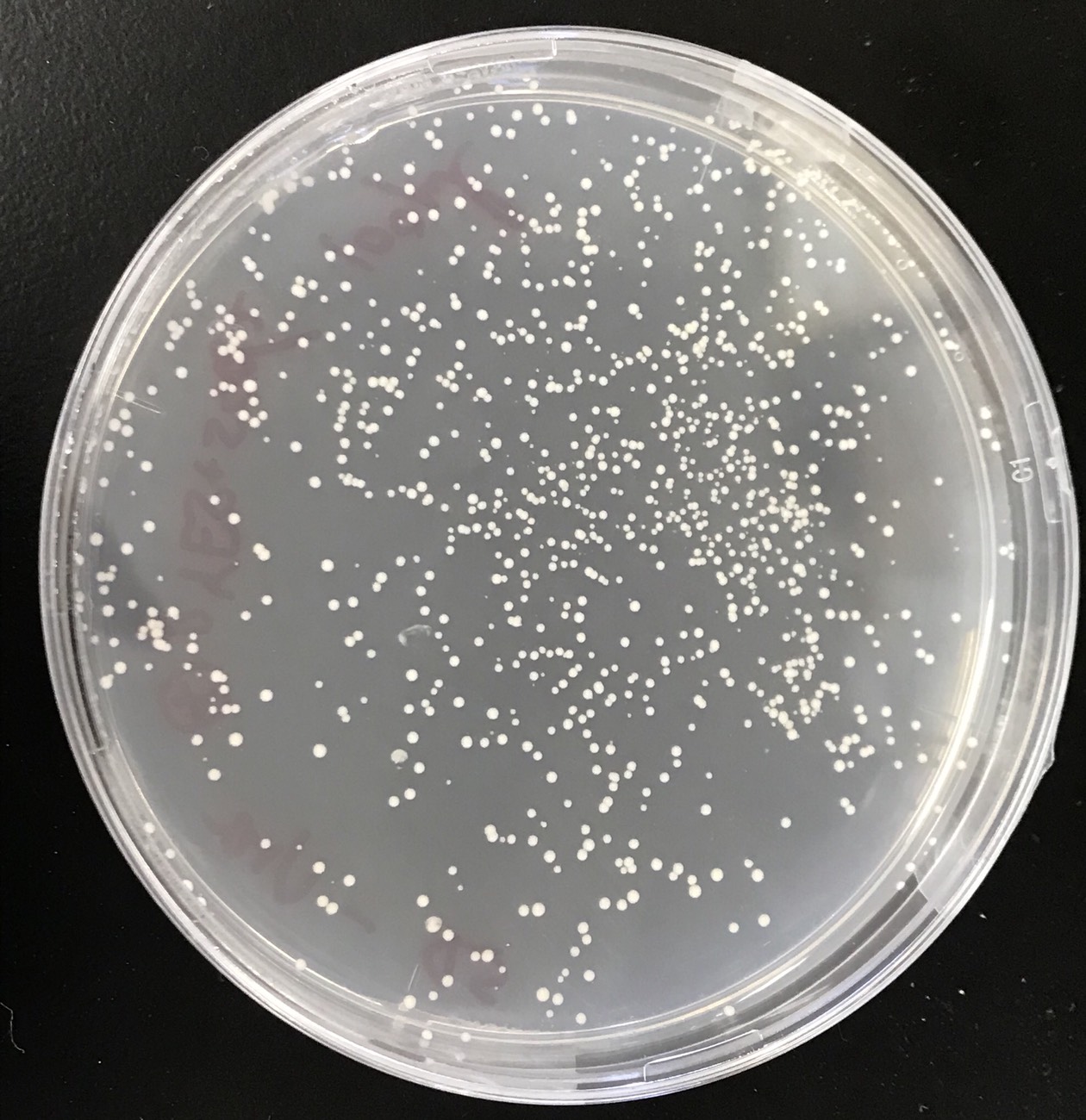
S. cerevisiae transformed was grown onto sd-ura medium. This figure(fig.6) shows that the pYES2 plasmid inserted sod2 was introduced into S. cerevisiae.
Silver
Confirmation that S. cerevisiae expressing sod2 acquired salt tolerance
Our project is done not by E. coli but by yeast.
Therefore, expression confirmation is carried out using plasmid for yeast(pYES2) instead of pSB1C3.
So, the submitted new part that checked expression for silver medal is not devices, but coding parts.
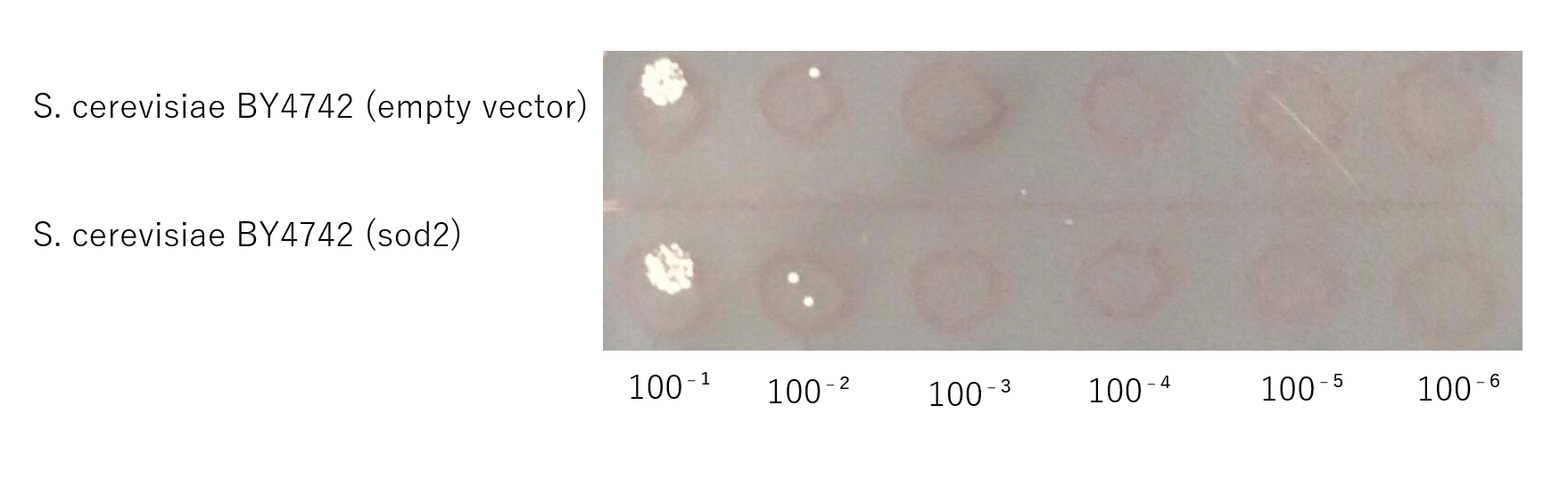
The growth of S. cerevisiae BY4742 cells expressing sod2 was tested on plates containing sodium.
This figure (fig.7) shows that S. cerevisiae cells that overexpress the sod2 product is more survived on SD-ura medium supplemented with 4.0 % (w/v) NaCl than the counterpart control S. cerevisiae transformed with an empty vector.